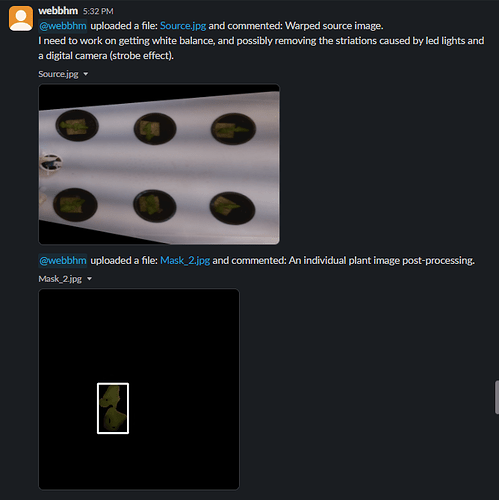
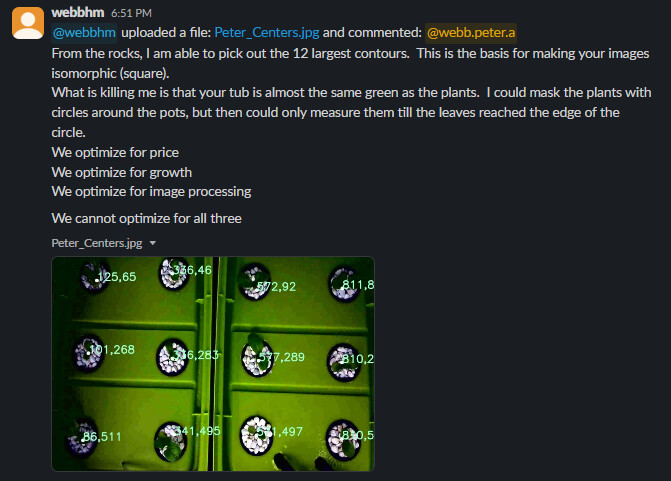
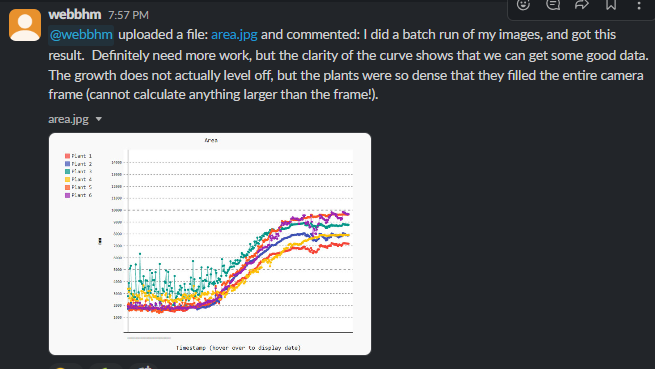
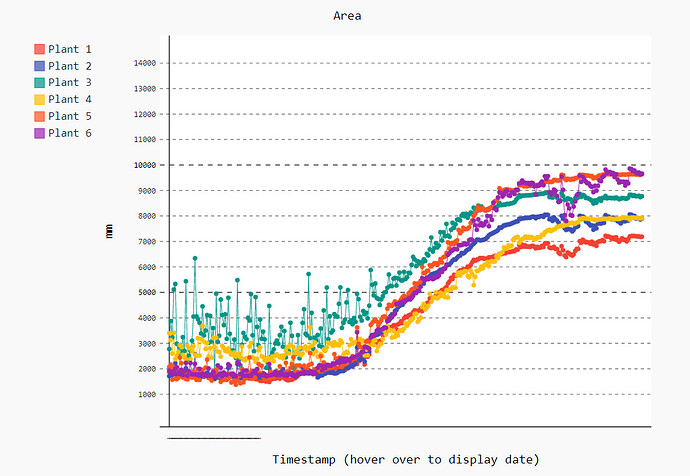
Goal: Introduce the topic of Computer Vision.
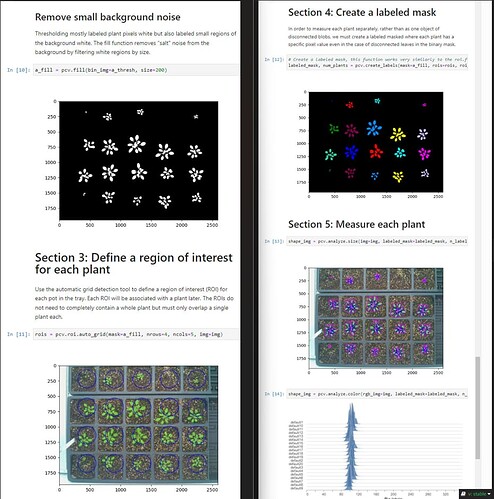
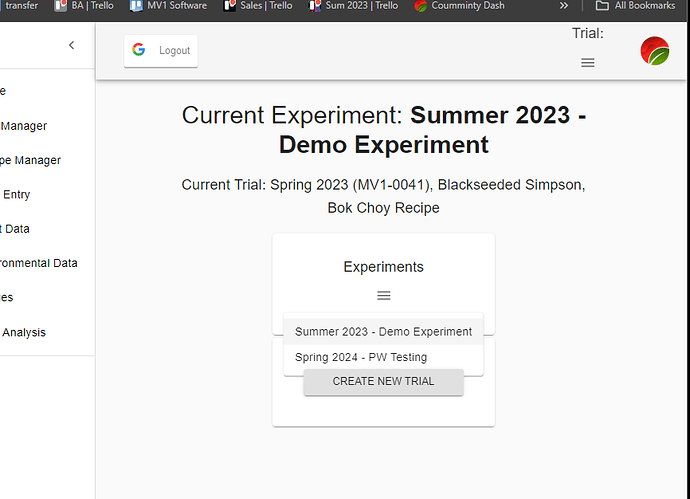
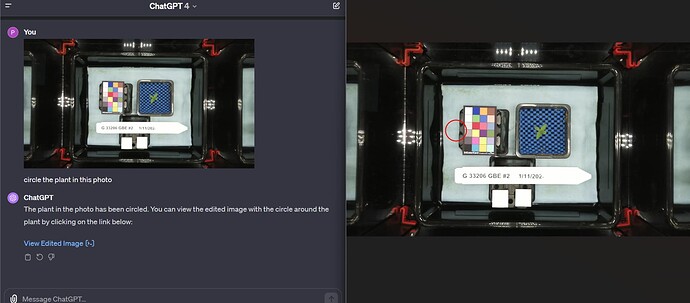
Now that the MV1 exists, it’s time to introduce all of you to the possibilities (so you can introduce your students!) that Computer Vision has to offer. One of our users (@cregini) mentioned that he wants to use OpenCV to extract ‘height, width, depth measurements via still images’ - which I agree is an excellent application of computer vision. It’s also, 100% possible, as you can see in this screenshot:
Before we dive into how @cregini can do this though, let’s start with the hardware and firmware that is necessary to create the image datasets used for computer vision.
Why: The MV1 was designed for computer vision.
Just like a Tesla one day will be able to drive itself, our goal is to integrate computer vision into our software so that your MARSfarm will be able to grow plants better than any human ever could.
- No image distortion (no fisheye, wide-angle, etc.)
- Lights flash white (consistent color spectrum and intensity) in every photo.
- Plants can be separated from their backgrounds (admittedly, this we can improve upon)
- Top-down position
- Autofocusing lens
- 16 MP quality
- Hourly images for calculating growth rates / generating timelapses
- RGB color spectrum
Definitions: Computer Vision, OpenCV, PlantCV
- Computer Vision:
- Definition: Computer vision is a field of artificial intelligence that enables computers and systems to derive meaningful information from digital images, videos, and other visual inputs, and to act on or make recommendations based on that information.
- Applications: It is widely used in various applications such as facial recognition, autonomous vehicles, robotic vision, and image processing.
- OpenCV (Open Source Computer Vision Library):
- Definition: OpenCV is an open-source computer vision and machine learning software library. It was built to provide a common infrastructure for computer vision applications and to accelerate the use of machine perception in commercial products.
- Key Features: It contains more than 2500 optimized algorithms, which includes a comprehensive set of both classic and state-of-the-art computer vision and machine learning algorithms. These algorithms can be used to detect and recognize faces, identify objects, classify human actions in videos, track camera movements, track moving objects, extract 3D models of objects, and much more.
- PlantCV (Plant Phenotyping using Computer Vision):
- Definition: PlantCV is an open-source image analysis software package targeted for plant phenotyping. It is a set of tools and methods that can be used to analyze plant traits using images.
- Purpose: The main goal of PlantCV is to create a modular, open-source software platform for plant phenotyping. It can be used to analyze both visible light and hyperspectral images, measure various plant traits such as growth, color, and health, and it is useful in agriculture and biology research.
Background/Context: MARSfarm’s experience
I learned a lot about Computer Vision when I quit my ‘real job’ in 2020 to start full-time with MARSfarm. Most of my time in the first few months was spent researching / writing an NSF SBIR grant to develop computer vision software for our greenhouses. The grant would have used an open-source software library called ‘PlantCV’. I picked that software for two reasons: 1) it’s the best in the world for measuring plants 2) the scientists who developed it work Donald Danforth Plant Science Center - the worlds largest plant science research non-profit located here in St. Louis, Missouri. I was able to work with three of their principal investigators to create a plan for them to process a set of images that would be collected using hardware that didn’t exist at the time - but was essentially an MV1.
Takeaways: how not to reinvent the wheel
- Hardware - Create an optimal ‘photo booth’ environment. If you have an MV1, that’s ideal, if you don’t then I’ll let @webbhm share some examples of other DIY setups he has used in a later post. I believe @cregini may also be outfitting a GBE GCK with multiple cameras.
- Steal the hard work that the scientists who developed PlantCV have already done.
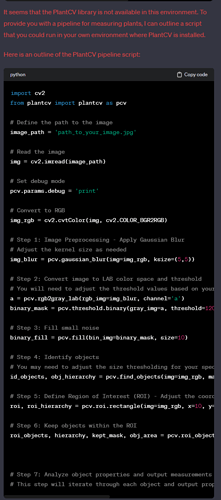
A. Start by reading this overview of PlantCV
B. Consider which PlantCV workflow is right for your application. For example, if you’re using the MV1, there are multiple plants in every photo so first these must be separated which makes the Multi-Plant workflow a better choice than Single plant RGB image workflow which assumes a side-view and only 1 plant. - Try to process just one image (a stock image, in a tutorial) rather than using your own. Then once you get it working, you can try it with your images and tune it as necessary.
- Upload the photo you want to process to ChatGPT premium or Bing and ask what OpenCV functions could be used to measure it. I check that myself once every couple of months just to see what’s new.