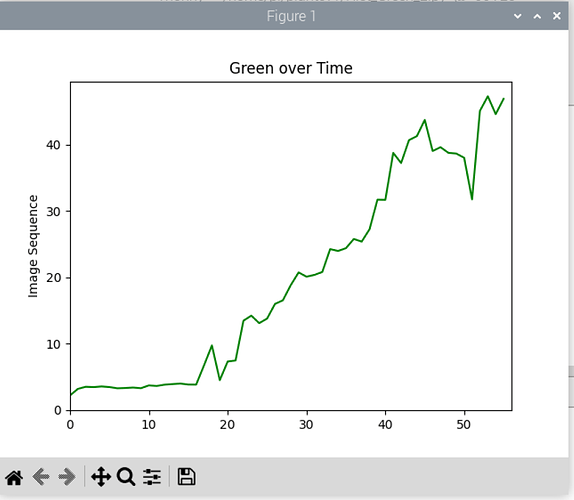
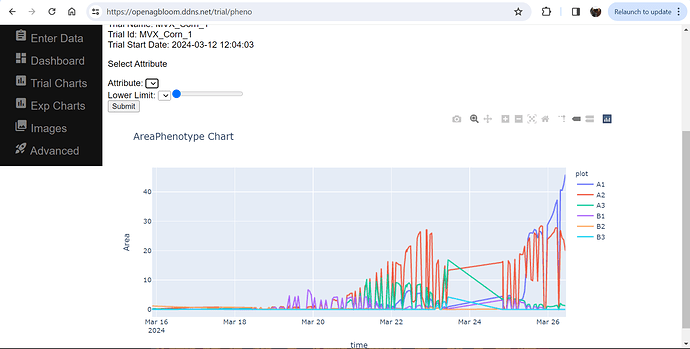
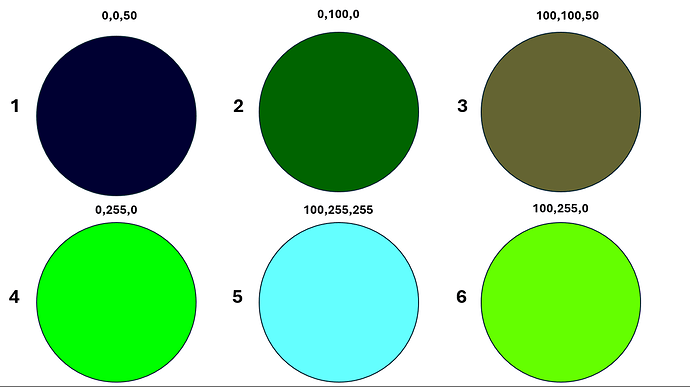
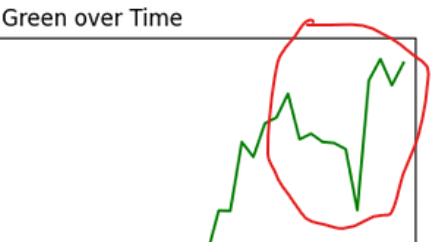
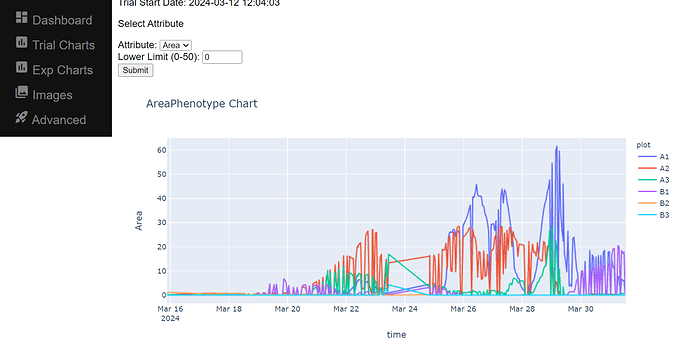
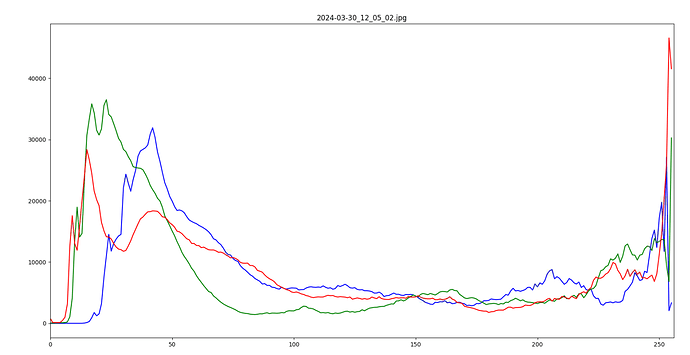
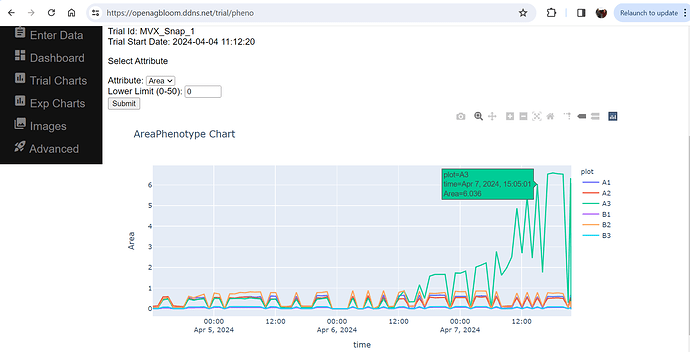
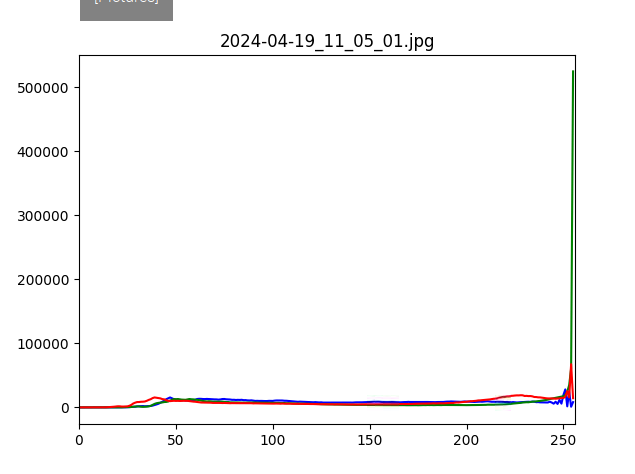
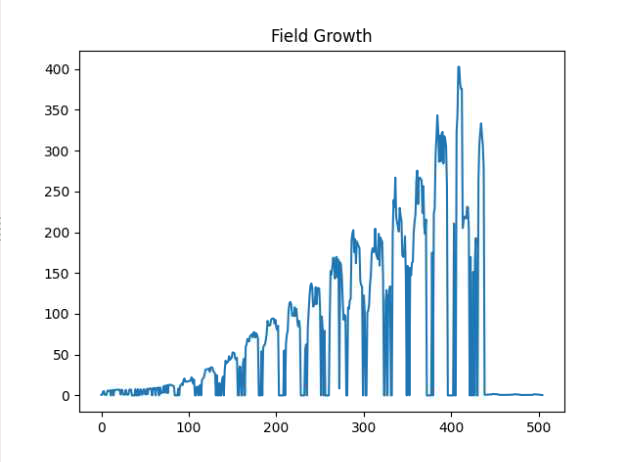
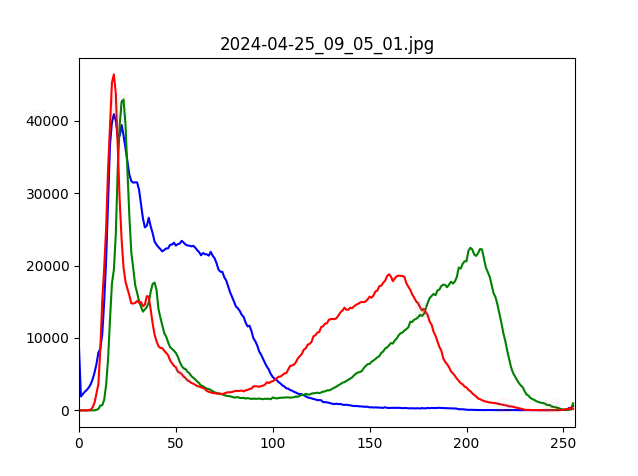
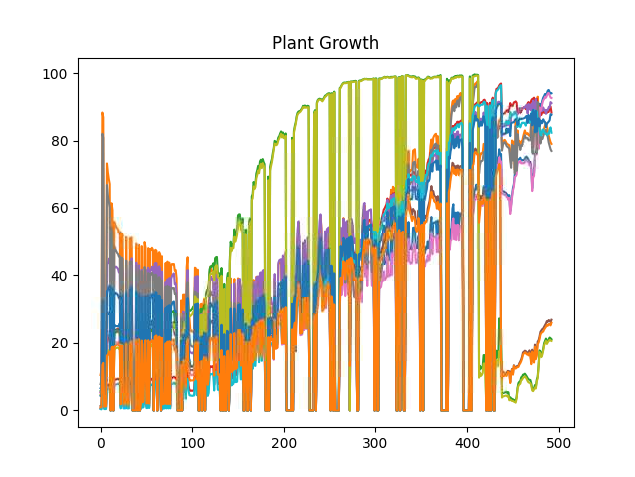
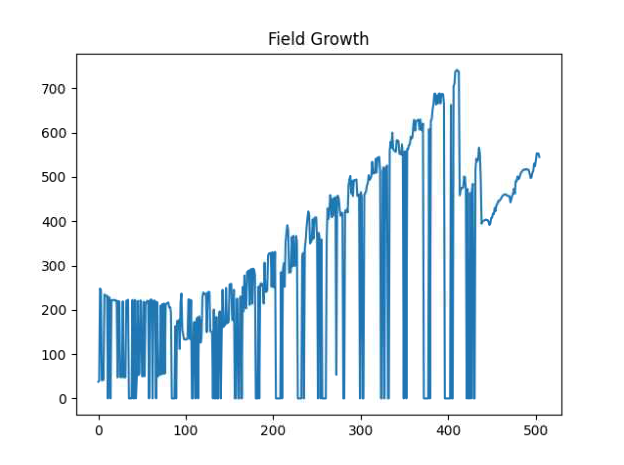
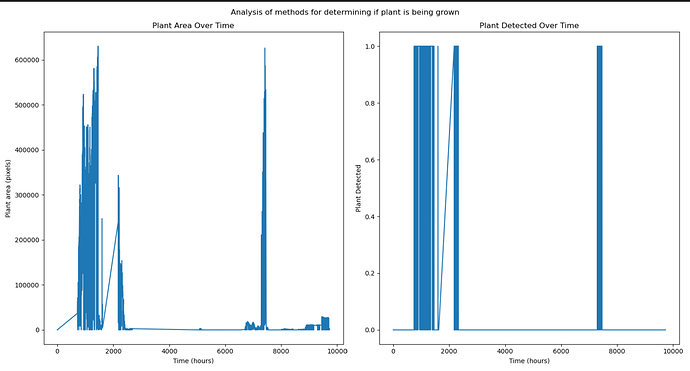
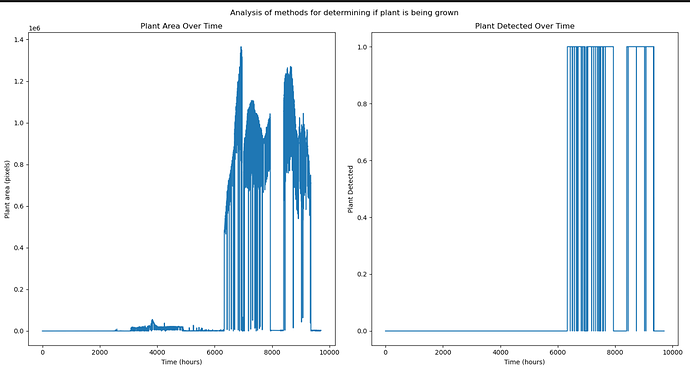
If you have PlantCV installed, it installs OpenCV as well, so the following code should work in your environment. Either use the attached file, or an image of your choice.
This is the file Find_Plant.py - the main file:
Test code working on a un-warped single image
import numpy as np
import cv2
import PlantUtil as fpu
#
v2.imread(pic)
#warp=fpu.transform(src, pts_in, pts_out)
#cv2.imshow('Warp', warp)
mask=fpu.getMask(src, lower, upper)
cv2.imshow('Mask',mask)
cv2.waitKey(2)
#res = cv2.bitwise_and(warp,warp, mask= mask)
edge=fpu.getEdges(mask)
cv2.imshow('Edges', edge)
cv2.waitKey(2)
ellipse=fpu.getContours(edge, src)
print(type(ellipse))
cv2.imshow('Elipse', ellipse)
#cv2.imwrite('/home/pi/MVP/Pictures/Ellipse.png', src2)
# Wat for the escape key to be pressed to end - does not work with Python IDE
k = cv2.waitKey(0) & 0xFF
cv2.destroyAllWindows()
PlantUtil.py are the utility functions that do the work:
# Routines to find plants and draw ellipse
import numpy as np
import cv2
import copy
def transform(src, pts_in, pts_out):
# Prepare the image for work, straighten and clip
#gimg=cv2.imread(pic, cv2.IMREAD_GRAYSCALE)
# Perform transformation warp
M=cv2.getPerspectiveTransform(pts_in, pts_out)
#print(M)
warp=cv2.warpPerspective(src, M,(724,720))
return warp
def getMask(img, lower, upper):
wh, bk, mask = get_pixels(img, upper, lower)
# erode and dialate the image to remove artifacts
kernel = np.ones((5,5), np.uint8)
erosion = cv2.erode(mask, kernel, iterations = 1)
dilation = cv2.dilate(erosion, kernel, iterations=5)
erosion2 = cv2.erode(dilation, kernel, iterations = 3)
# Create a test image showing the green through the mask
# Not actually used, but interesting to play with
return erosion2
#bgr=cv2.cvtColor(erosion2, cv2.COLOR_HSV2BGR)
#gr=cv2.cvtColor(res, cv2.COLOR_BGR2GRAY)
def get_pixels(img, upper, lower):
#Begin finding of plants
# Convert BGR to HSV
hsv = cv2.cvtColor(img, cv2.COLOR_BGR2HSV)
#print("image type: ", type(hsv))
# Threshold the HSV image to get only green colors and create mask of greens
mask = cv2.inRange(hsv, lower, upper)
height, width = mask.shape[:2]
wh = np.sum(mask == 255)
bk = np.sum(mask == 0)
sz = wh + bk
return wh, bk, mask
#print(f'Size: {height * width}')
#print(f'Mask Size: {sz}')
print(f'White: {wh}')
print(f'Black: {bk}')
print(f'Ratio: {round(wh/sz, 2)}')
def getEdges(img):
BLACK=[0, 0, 0]
#Border the image so edges run around ends
bdr=cv2.copyMakeBorder(img, 10, 10, 10, 10, cv2.BORDER_CONSTANT, value=BLACK)
# Detect the edges of the plants
edges=cv2.Canny(bdr, 100, 200)
print(edges.shape)
return edges
def getContours(img, display):
# Build contours of the plants from the edges
# Only get top level (external) contours
contours,hierarchy= cv2.findContours(img, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
# Copy the image, otherwise it would be overwritten and no longer available
out=copy.copy(display)
# Walk all the contours
x=0 # Counter for display
for i in contours:
x=x+1
# Create and draw the elipses - this is the final output image
if cv2.contourArea(i) > 5:
ellipse = cv2.fitEllipse(i)
cv2.ellipse(out, ellipse,(0,0,0),2)
else:
print("Too small of area for evaluation", x, cv2.contourArea(i))
# The area could be used to calculate phenotype info
# print("Contour: ", x, "Area: ", cv2.contourArea(i))
return out
''''